A molecular control hub maintains order
How are proteins in our cells modified while they are still being synthesized? An international team of researchers from the University of Konstanz, Caltech, and ETH Zurich has deciphered the molecular mechanism of this vital process.
Proteins are among the most important molecular building blocks of life. They are formed from amino acids that are chained together based on the information in our genetic material. In this process, the code of our genes is translated into a sequence of amino acids. However, this translation is only the first step. Often, special enzymes modify the proteins while they are leaving their cellular production site – the ribosome. Only after this can the proteins fulfil their diverse biological functions.
What was previously poorly understood: How do these enzymes work together to modify proteins at the ribosome? And how is their activity regulated and coordinated? An international team of researchers from the University of Konstanz (Germany), the California Institute of Technology (Caltech; USA) and ETH Zurich (Switzerland) has now unravelled the complex molecular mechanism for two consecutive protein modifications, which in mammals affect about 40 percent of all proteins. This vital process is coordinated by a protein complex called NAC (short for "nascent polypeptide-associated complex"). The results have recently been published in the journal Nature.
Essential for normal cell function
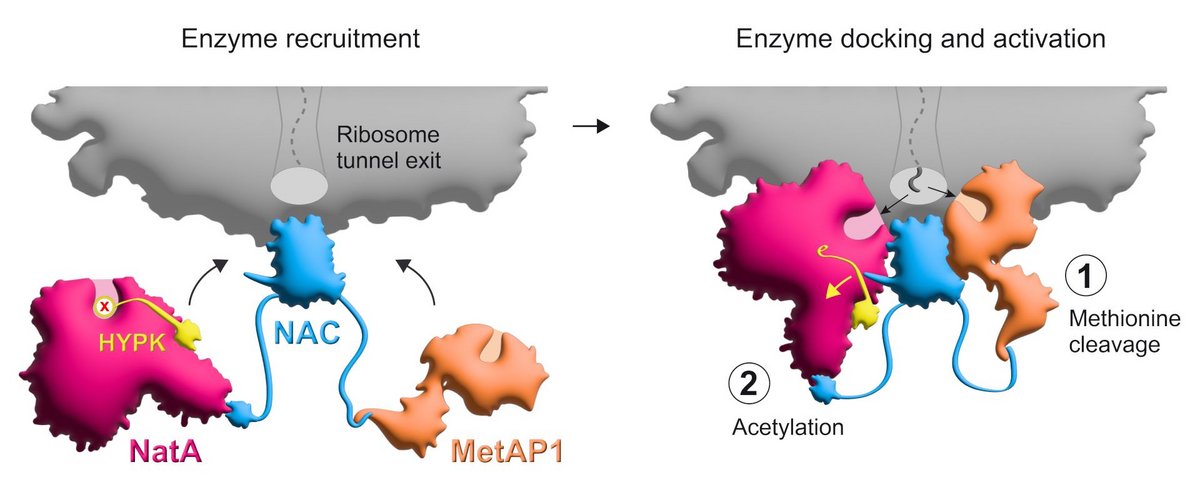
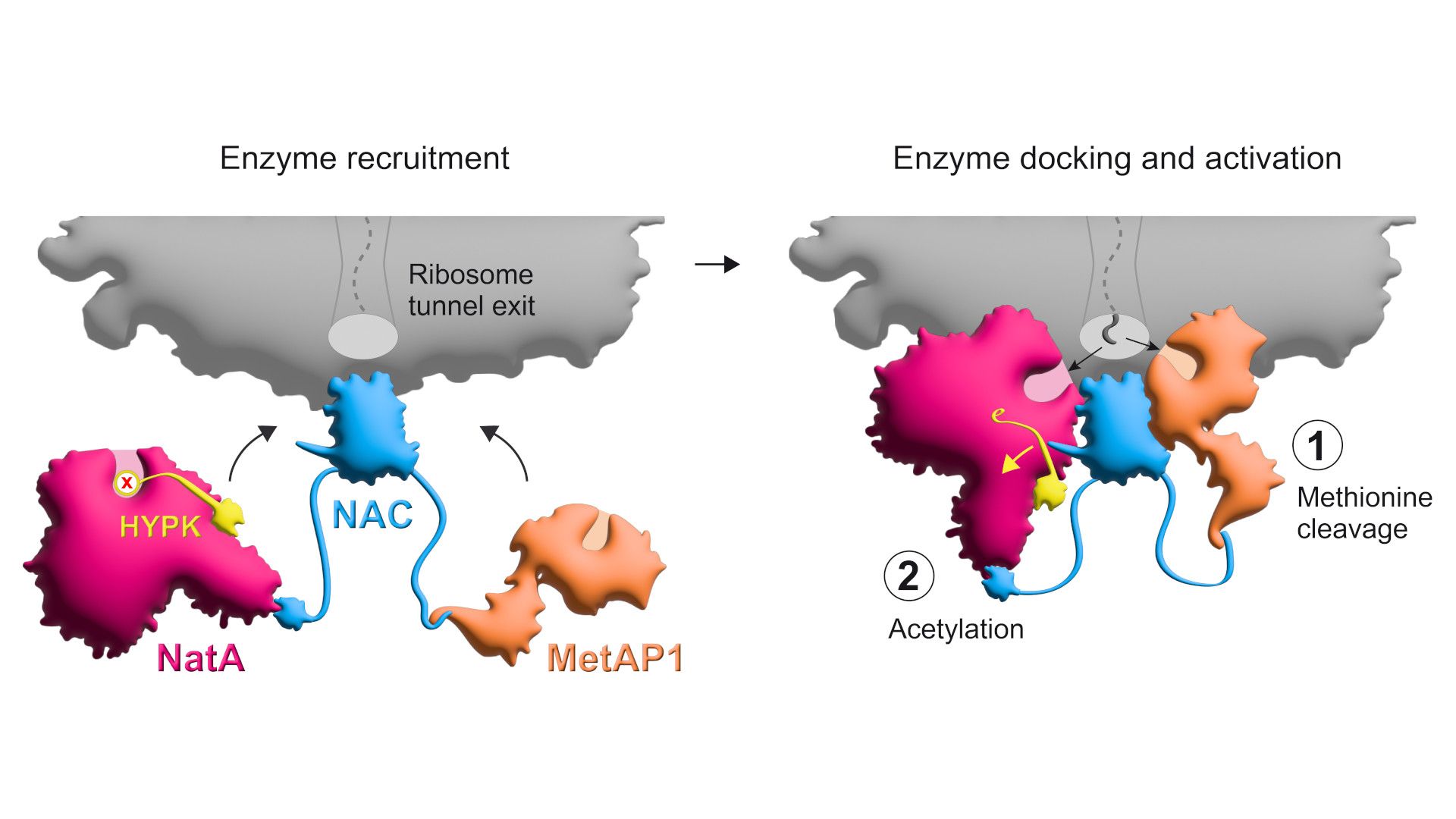
More specifically, these modifications are the removal of the amino acid methionine from the growing protein, followed by the attachment of what is known as an acetyl group to the remaining end. Both of these processes take place at the ribosomal tunnel, i.e. at the location where the proteins leave the ribosome as a growing chain of amino acids during synthesis. These modifications are absolutely essential for the majority of our proteins, as they influence several protein properties at the same time – such as their three-dimensional folding, lifespan or interaction with other proteins – and thus the proper function of the proteins.
"A dysregulation of the processes involved in protein modification can have extremely negative consequences for the organism. It is associated with developmental disorders or diseases such as cancer and Parkinson's, for example," explains Elke Deuerling, professor at the University of Konstanz’s Department of Biology. With her colleagues Martin Gamerdinger and Laurenz Rabl from Konstanz she significantly contributed to the current study.
The mechanism in detail
The time slot for the cleavage of methionine and the subsequent acetylation to occur smoothly is quite short. During this time, several enzymes must be brought to the right place and be regulated: MetAP1, which causes the cleavage of methionine, and NatA for the subsequent acetylation. However, NatA is normally bound by an inhibitor protein, HYPK, which suppresses its function. By combining biochemical, structural and in vivo experiments, the Konstanz researchers and their colleagues from the USA and Switzerland have now succeeded in shedding light on how this complex process is controlled and how the macromolecules involved interact.
The protein complex NAC plays a central role in this process: NAC is located at the exit of the ribosomal tunnel, where newly synthesized proteins emerge. From there, it recruits the enzymes required for the cleavage of methionine and the subsequent acetylation and positions them with their biochemically active regions at the appropriate locations near the tunnel to access the nascent protein. "In addition, NAC induces NatA to lose its inhibitory contact with HYPK. This ensures that NatA's function is only activated at the ribosome, where it can then mediate the desired acetylation,” says Shu-ou Shan, the Altair Professor of Chemistry and executive officer for biochemistry and molecular biophysics at Caltech.
NAC: a general molecular control hub?
It is known from previous studies that NAC also recruits other factors to the ribosomal tunnel besides the enzymes MetAP1 and NatA. "We assume that NAC has the function of an even more elaborate molecular control hub. It ensures that the nascent proteins have access to different components of the cell's biochemical toolkit as they leave the ribosome, depending on the requirements," explains Gamerdinger.
The current study shows how NAC fulfils this important function in the specific case of methionine cleavage followed by an acetylation. This new knowledge about the complex molecular processes in our cells allows us to better understand the biochemical foundations of life. In addition, it will contribute to a better understanding of how dysregulations of the components involved in the modification of proteins lead to the development of diseases. In the long term, this could function as a basis for the development of new therapeutic approaches in medicine.
Key facts:
- Original publication: A. Lentzsch, D. Yudin, M. Gamerdinger, S. Chandrasekar, L. Rabl, A. Scaiola, E. Deuerling, N. Ban & S. Shan (2024) NAC guides a ribosomal multienzyme complex for nascent protein processing. Nature; doi: 10.1038/s41586-024-07846-7
- An international research team involving researchers from the University of Konstanz, the California Institute of Technology, and ETH Zurich has unravelled a molecular mechanism of cotranslational protein modification in eukaryotes.
- Researchers from the Department of Biology at the University of Konstanz who were involved in the study: Prof. Elke Deuerling, Dr. Martin Gamerdinger and Laurenz Rabl
- Funding: National Institutes of Health (NIH), National Science Foundation (NSF), German Research Foundation (DFG), Swiss National Science Foundation (SNSF), and European Research Council